I found a really good code at GitHub for fitting a Gaussian Mixture Model (GMM) with Expectation Maximization (EM) for ROS. There are so many parameters that you can change. Some of the most important ones are:
|
1 2 3 4 5 6 7 |
// minimum number of gaussians #define PARAM_NAME_GAUSSIAN_COUNT_MIN "gaussian_count_min" #define PARAM_DEFAULT_GAUSSIAN_COUNT_MIN 1 // search will terminate when the gaussian count reaches this #define PARAM_NAME_GAUSSIAN_COUNT_MAX "gaussian_count_max" #define PARAM_DEFAULT_GAUSSIAN_COUNT_MAX 10 |
To find the optimal number of components, it uses Bayesian information criterion (BIC). There are other methods to find the optimal number of components: Minimum description length (MDL), Akaike information criterion (AIC), Minimum message length (MML).
Here is my code for generating a 2 Gaussian and sending them to this node:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 |
Eigen::Vector3f mean; Eigen::Matrix3f covar; float theta; mean << 0,0,0; // Set the mean /* first covar matrix: |10 0 0| |0 1 0| |0 0 1| */ covar <<10,0,0,0,1,0,0,0,1; //first gaussian Eigen::EigenMultivariateNormal<float> normX_solver1(mean,covar); first_gaussian_data_eigen<< normX_solver1.samples(n_rows).transpose() ; //second gaussian mean << 4,3,5; // Set the mean /* second covar matrix: |1 0 0| |0 1 0| |0 0 1| */ covar <<1,0,0,0,1,0,0,0,1; Eigen::EigenMultivariateNormal<float> normX_solver2(mean,covar); second_gaussian_data_eigen<<normX_solver2.samples(n_rows).transpose() ; //concat two gaussians total_gaussian_data_eigen<<first_gaussian_data_eigen,second_gaussian_data_eigen; |
and you need to put them in to send them to the node:
|
1 2 3 4 5 6 7 8 9 10 11 |
std_msgs::Float32MultiArray trajectory_points; trajectory_points.layout.dim.resize(2); // The first dimension of the layout (at position 0) has size equal to the number of samples (number of rows). trajectory_points.layout.dim[0].stride = total_gaussian_data_eigen.innerSize(); trajectory_points.layout.dim[0].size = total_gaussian_data_eigen.innerSize(); // The second dimension of the layout has size equal to the number of elements in each tuple (number of columns). trajectory_points.layout.dim[1].stride = total_gaussian_data_eigen.size(); trajectory_points.layout.dim[1].size = total_gaussian_data_eigen.outerSize(); |
and the results are what we expect:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 |
[ INFO] [1510838448.806649183]: gmm: attempting gaussian count: 1 [ INFO] [1510838448.871223668]: gmm: bic is: 2533.130859 [ INFO] [1510838448.871256190]: gmm: attempting gaussian count: 2 [ INFO] [1510838449.432518373]: gmm: bic is: 2273.819580 [ INFO] [1510838449.432552831]: gmm: attempting gaussian count: 3 [ INFO] [1510838453.088420451]: gmm: bic is: 2295.604248 [ INFO] [1510838453.088457743]: gmm: chosen gaussian count 2 [ INFO] [1510838453.088472330]: weight 0.499777: [ INFO] [1510838453.088481566]: means are: [ INFO] [1510838453.088492342]: -0.137626 [ INFO] [1510838453.088501603]: -0.008509 [ INFO] [1510838453.088508936]: -0.009013 [ INFO] [1510838453.088520380]: weight 0.500223: [ INFO] [1510838453.088530897]: means are: [ INFO] [1510838453.088539627]: 3.991702 [ INFO] [1510838453.088548201]: 2.990528 [ INFO] [1510838453.088556485]: 4.955379 |
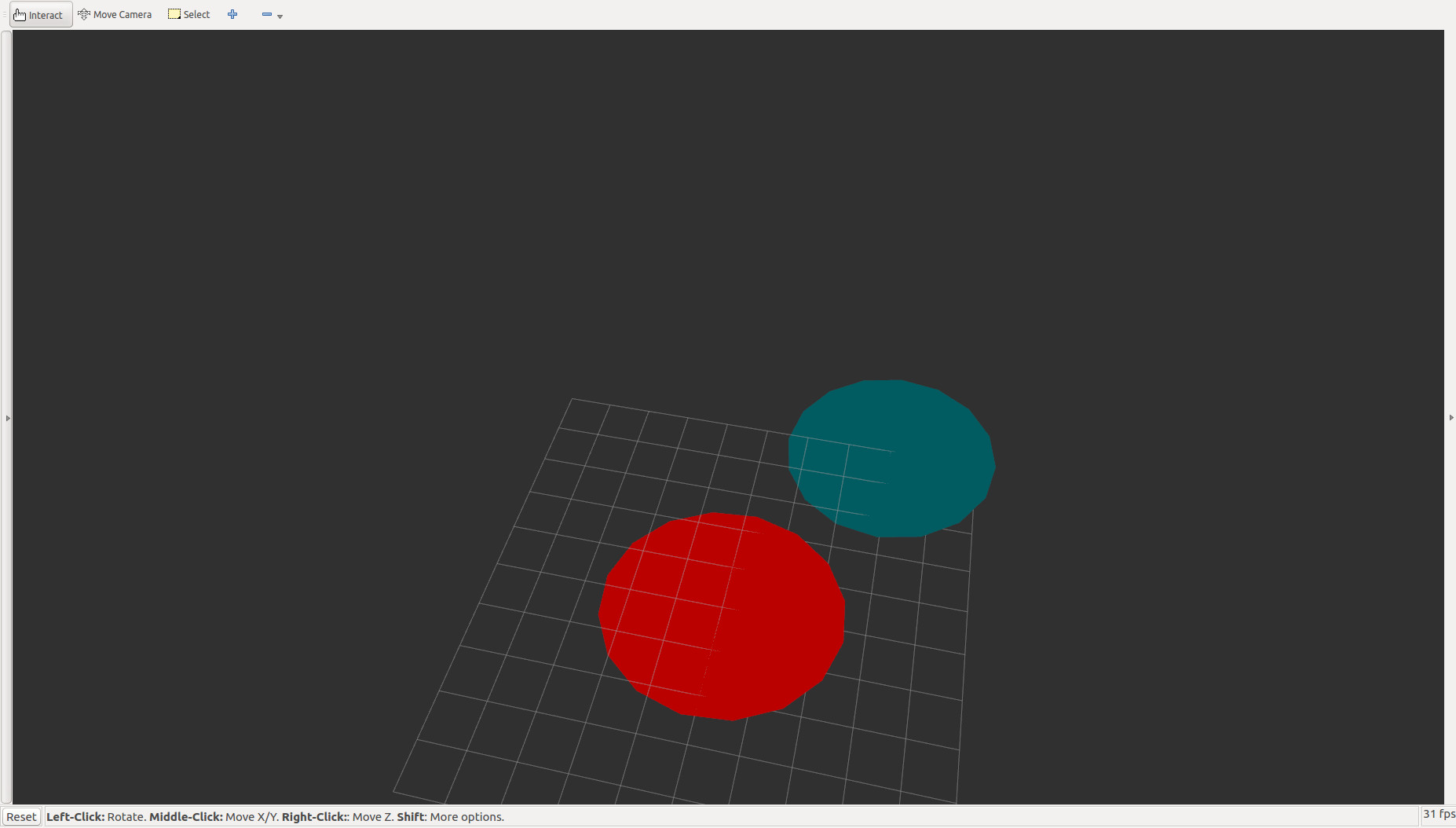
It also makes it possible to visualize the data in RVIZ, but first, you have to publish your tf data and set the frame name and topic names correctly in gmm_rviz_converter.h
|
1 2 3 4 5 6 7 8 9 10 11 |
#define PARAM_NAME_INPUT_TOPIC "input_topic" //#define PARAM_DEFAULT_INPUT_TOPIC "/gmm_rviz_converter_input" #define PARAM_DEFAULT_INPUT_TOPIC "/gmm/mix" #define PARAM_NAME_OUTPUT_TOPIC "output_topic" #define PARAM_DEFAULT_OUTPUT_TOPIC "/gmm_rviz_converter_output" #define PARAM_NAME_FRAME_ID "frame_id" //#define PARAM_DEFAULT_FRAME_ID "/root" #define PARAM_DEFAULT_FRAME_ID "/base_link" |
and add a MarkerArray in RVIZ and set the topic “gmm_rviz_converter_output”